Step-by-step tutorial
This tutorial introduces the foundations of the QMENTA SDK and the technological ecosystem that surrounds it, including Docker and the QMENTA Platform. Its main objective is to guide you through the complete lifecycle of a QMENTA tool: from local implementation and testing, to deployment and execution within the QMENTA Platform.
In QMENTA, a tool is a self-contained piece of software—typically packaged as a Docker image—that performs a well-defined computational task on medical data. Tools define their expected inputs, outputs, and execution logic, and can be run reproducibly at scale through the QMENTA Platform. By the end of this tutorial, you will understand how tools are structured, how they interact with the QMENTA Platform, and how to validate that they work correctly before making them available to end users.
The final result of this tutorial, including the complete implementation, can be found in this GitHub repository.
As a practical example, we will adapt the ANTs Tutorial, originally implemented as a Python notebook, into a QMENTA tool. The example will use a pair of T1-weighted MRI brain slices as input data. The tool will expose several configurable input settings. In the QMENTA Platform interface the will allow the user to select which ANTs processing steps to execute:
Perform bias field correction
Run tissue segmentation
Compute cortical thickness
Register two images using
The tissue segmentation step must be executed before cortical thickness estimation, as the latter depends on the segmentation results. For this reason, these two steps are linked within the workflow. All other steps are independent and can be run separately.
In addition, the tool provides a dedicated setting that allows users to fine-tune the analysis by adjusting a specific segmentation parameter.
The resulting tool will extract some statistics, segment anatomical neuroimaging data (2D slice) and report the segmentation results in the online viewer, demonstrating how an existing analysis workflow can be transformed into a reusable and deployable QMENTA tool. In case you are curious, you can check what kind of tools QMENTA has already available in the QMENTA Tools Catalog.

Note
Advanced Normalization Tools (ANTs) is a C++ library available through the command line that computes high-dimensional mappings to capture the statistics of brain structure and function. It allows one to organize, visualize and statistically explore large biomedical image sets. Additionally, it integrates imaging modalities in space + time and works across species or organ systems with minimal customization.
Before you begin
To complete this tutorial, you will need the following:
Your favorite IDE or text editor.
Basic knowledge of Python and its ecosystem.
A QMENTA Platform account. You can create a new account in this registration page.
Docker installed in your system. Check their getting started guide to know more about that.
A DockerHub account to register your image.
The example data provided here: a T1 of a segmentated brain r16, and a T1 of a segmentated brain r64.
A python>=3.10 environment with the qmenta-sdk-lib>=2.0 and antspyx>=0.6.1 libraries installed.
Getting your processing tool in the QMENTA Platform involves several well-defined steps that transform a local script into a reusable, executable component of the QMENTA Platform. At a high level, the process includes the following stages:
Implement the tool logic by writing the main Python script that performs the desired computation and handles inputs and outputs according to the tool specification.
Define the execution environment by creating a Docker image that contains all required dependencies, libraries, and system configurations needed for the tool to run reproducibly.
Test the tool locally to ensure that the code and Docker container behave as expected before deployment.
Register and deploy the tool in the QMENTA Platform.
Run the command tool_maker and it should launch the graphical user interface of the Tool Maker.
/home/myfolder$ tool-maker
It will show something similar to the following screenshot:
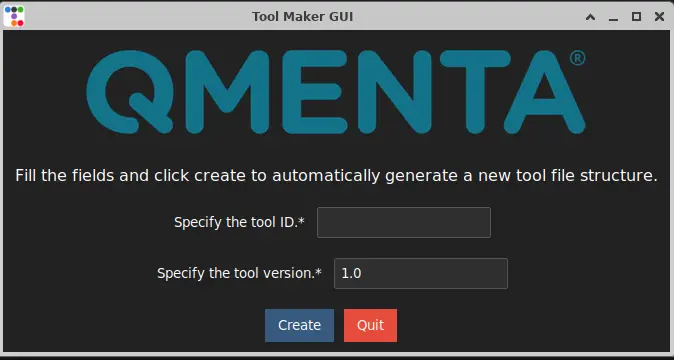
Enter a tool ID (it only accepts lower case letters and “_” character) For example: tool_maker_tutorial. Click on “Create” to create the basic tool file structure.
Generated Files:
/home/myfolder/local_tools/
└─ my_first_tool
├─ description.html
├─ local
│ ├─ Dockerfile
│ ├─ requirements.txt
│ └─ test
│ ├─ sample_data
│ └─ test_tool.py
└─ tool.py
Open you code editor inside the created local_tools/tool_maker_tutorial folder.
Write the main Python program
Input definition and configuration
Open the file run.py. You can see the basic imports are already created and the class for the tool is written. We are going to use ants as well so make sure you import it in the top of the file:
import ants
Let’s start editing the method tools_input().
These settings define the minimum requirements and configuration options that must be satisfied before the tool can start processing. They describe which input data is required, how that data is selected, and which processing steps will be executed. Together, they form the user-facing interface of the tool in the QMENTA Platform and ensure that the analysis is configured correctly and consistently.
First, an informational message is displayed to the user, providing a brief description of the tool and its purpose.
The input container defines the medical images required by the tool. In this case, the tool expects anatomical brain images with accepted modalities T1 or T2. Two separate input files are required:
One image tagged with r16
One image tagged with r64
Each input is mandatory, and exactly one file must be provided for each condition. The use of file_filter_condition_name allows the tool to distinguish between these inputs internally and apply different processing logic if needed.
Visual separators (horizontal lines and headings) are then used to improve readability and structure in the tool configuration interface.
Next, a multiple-choice input allows the user to select which processing steps to execute. These options correspond to the main stages of the ANTs-based workflow, such as bias field correction, tissue segmentation, cortical thickness estimation, and image registration. Default selections are provided to guide typical usage while still allowing flexibility.
Finally, a string input parameter is used to configure the MRF (Markov Random Field) parameters required by the segmentation step. This parameter controls the amount of spatial smoothing and the neighborhood radius used during processing. The value is provided as a string to match the ANTs-style parameter format, making it easy to adapt the tool to different image dimensions or analysis requirements.
Together, these settings ensure that all necessary inputs and parameters are defined before execution, enabling the tool to run reliably and reproducibly on the QMENTA Platform.
In the auto-generated code you can find a useful link to the different inputs’ definition and configuration, helpful if you have not checked it yet to understand better these lines of code.
Replace the content of the tools_input() with this:
# Displays a text box in blue color with the information specified.
self.add_info(
"ANTsPy tutorial settings." \
)
# Add the file selection method:
self.add_input_container(
title="Oncology medical image",
info="Required inputs:<br><b>• Anatomical brain medical image</b>: 2D image to analyze<br> Accepted modalities: 'T1', 'T2'", # text in case we want to show which modalities are accepted by the filter.
anchor=1,
batch=1,
container_id="input_images",
mandatory=1,
file_list=[
InputFile(
file_filter_condition_name="c_image1",
filter_file=FilterFile(
modality=[Modality.T1, Modality.T2],
tags=[["r16"]]
),
mandatory=1,
min_files=1,
max_files=1,
),
InputFile(
file_filter_condition_name="c_image2",
filter_file=FilterFile(
modality=[Modality.T1, Modality.T2],
tags=[["r64"]]
),
mandatory=1,
min_files=1,
max_files=1,
),
],
)
# Displays an horizontal line
self.add_line()
# Displays a header text
self.add_heading("ANTs tool can perform the following steps")
self.add_input_multiple_choice(
id_="perform_steps",
options=[
["do_biasfieldcorrection", "Perform Bias Field Correction"],
["do_segmentation", "Perform tissue segmentation"],
["do_thickness", "Perform cortical thickness"],
["do_registration", "ANTs registration interface"],
],
default=["do_biasfieldcorrection", "do_segmentation"],
title="Which steps do you want to execute?",
)
# Displays an horizontal line
self.add_line()
# this parameter will modify the 'm' parameter in `atropos` method of ants (segmentation step)
self.add_input_string(
id_="mrf",
default="[0.2, 1x1]", # antspy tutorial uses 2D iamge
title="'mrf' parameters as a string, usually \"[smoothingFactor,radius]\" " \
"where smoothingFactor determines the amount of smoothing and radius determines " \
"the MRF neighborhood, as an ANTs style neighborhood vector eg \"1x1\" for a 2D image.",
)
We have defined an input container that will filter two files:
One file that can have modality T1 or T2, and the tag r16.
One file that can have modality T1 or T2, and the tag r64.
We have added an horizontal line to divide the settings into sections
Processing code (run)
Next, you will begin implementing the run()` function, which is the core entry point of the tool execution. This method is responsible for orchestrating the full data processing workflow: preparing the execution environment, retrieving input data, processing the data, and producing the final results.
As a first step, fetch and download the input data associated with the analysis. Input handling is managed through the QMENTA SDK, which provides utilities to automatically retrieve all inputs defined for the tool. By calling:
self.prepare_inputs(context, logger)
the SDK downloads the input files and stores them internally within the self.inputs object, making them readily available for use during processing.
In this tutorial, the tool uses a single input container named input_images. Within this container, the file_filter_condition_name argument is used to further refine which files are processed by the tool. This mechanism allows the tool to handle specific subsets of data when multiple files or modalities are present.
After preparing the inputs, you can access the downloaded files using the following pattern:
c_files_handlers_list = self.inputs.input_images.c_files
The resulting variable, c_files_handlers_list, is a list of file handler objects, where each element represents a single input file. These handlers expose methods and attributes that allow you to interact with the files, such as retrieving their local paths or associated metadata. A full list of available functionalities can be found in the SDK documentation.
To obtain the local filesystem path of a file—for example, the first file in the list—you can use:
input_data_path = c_files_handlers_list[0].file_path
To monitor the execution flow and facilitate debugging, you should also initialize a logger. Logging allows you to track the progress of the tool, record key events (such as when data is downloaded or processing starts), and capture error messages in a structured way. This is particularly important when tools are executed remotely on the QMENTA Platform, where logs are often the primary mechanism for understanding what happened during execution.
In our particular example, the first part of the run() method looks like this:
# ============================================================
# INITIAL SETUP
# ============================================================
logger = logging.getLogger("main")
logger.info("Tool starting")
# Define and create working directory
output_dir = os.path.join(os.environ.get("WORKDIR", "/root"), "OUTPUT")
os.makedirs(output_dir, exist_ok=True)
os.chdir(output_dir)
# this will report in the QMENTA Platform interface the starting of the tool
context.set_progress(value=0, message="Initializing tool execution")
# Download inputs and populate self.inputs
context.set_progress(message="Downloading input data")
self.prepare_inputs(context, logger)
# Retrieve input image paths
fname1_handler = self.inputs.input_images.c_image1[0]
fname1 = fname1_handler.file_path
# Second image only required if Registration is selected to be done.
fname2 = None
if "do_registration" in self.inputs.perform_steps:
fname2 = self.inputs.input_images.c_image2[0].file_path
More details about containers, file filters and other input settings can be found in Parameters and Input Files.
To keep track of the files generated by the tool and what needs to be reported we initialize two lists:
# Containers to track outputs
generated_files = []
report_items = []
The settings of the tool, that will be used at a later stage to control the execution of the ANTs library, are accessible from self.inputs followed by the setting id.
At the top of the tool script, before the class definition, you can include an HTML template that will be used to generate the report:
HTML_TEMPLATE = """
<!DOCTYPE html>
<html>
<head>
<meta charset="UTF-8">
<title>Image with Caption</title>
</head>
<body>
{body}
</body>
</html>
"""
BODY_TEMPLATE ="""
<h1>{header}</h1>
<h3>{image_caption}</h3>
<img src="{src_image}" alt="{image_description}" style="max-width: 400px;">
"""
Now it’s time for some heavy lifting: you will process the different steps, checking if the step was selected by the user:
# ============================================================
# PROCESSING PHASE
# ============================================================
logger.info("Starting processing phase")
logger.info("Selected steps:\n{}".format("\n".join(self.inputs.perform_steps)))
img = ants.image_read(fname1)
# --- Bias Field Correction ---
if "do_biasfieldcorrection" in self.inputs.perform_steps:
logger.info("Running N4 bias field correction")
image_n4 = ants.n4_bias_field_correction(img)
image_n4.to_filename("n4_processed.nii.gz")
generated_files.append("n4_processed.nii.gz")
report_items.append({
"header": "N4 Bias Field Correction",
"image": "n4_bias.png",
"source_image": image_n4,
"description": "N4 bias field correction result",
})
# Enforce dependency: segmentation required for thickness
if "do_thickness" in self.inputs.perform_steps and "do_segmentation" not in self.inputs.perform_steps:
self.inputs.perform_steps.append("do_segmentation")
# --- Tissue Segmentation ---
if "do_segmentation" in self.inputs.perform_steps:
logger.info("Running tissue segmentation")
mask = ants.get_mask(img)
img_seg = ants.atropos(
a=img,
m=self.inputs.mrf,
c='[2,0]',
i='kmeans[3]',
x=mask
)
img_seg["segmentation"].to_filename("atropos_processed.nii.gz")
generated_files.append("atropos_processed.nii.gz")
report_items.append({
"header": "Tissue Segmentation",
"image": "segmentation.png",
"source_image": img_seg["segmentation"],
"description": "ANTs Atropos tissue segmentation",
})
# --- Cortical Thickness ---
if "do_thickness" in self.inputs.perform_steps:
logger.info("Running cortical thickness estimation")
thickimg = ants.kelly_kapowski(
s=img_seg["segmentation"],
g=img_seg["probabilityimages"][1],
w=img_seg["probabilityimages"][2],
its=45, r=0.5, m=1
)
thickimg.to_filename("thickness_processed.nii.gz")
generated_files.append("thickness_processed.nii.gz")
report_items.append({
"header": "Cortical Thickness",
"image": "thickness.png",
"overlay": thickimg,
"base_image": img,
"description": "Cortical thickness estimation",
})
# --- Registration ---
if "do_registration" in self.inputs.perform_steps:
logger.info("Running image registration")
fixed = ants.image_read(fname1)
moving = ants.image_read(fname2)
tx = ants.registration(
fixed=fixed,
moving=moving,
type_of_transform="SyN"
)
warped = tx["warpedmovout"]
warped.to_filename("warped.nii.gz")
generated_files.append("warped.nii.gz")
report_items.append({
"header": "Image Registration",
"image": "registration.png",
"overlay": warped,
"base_image": fixed,
"description": "ANTs nonlinear registration result",
})
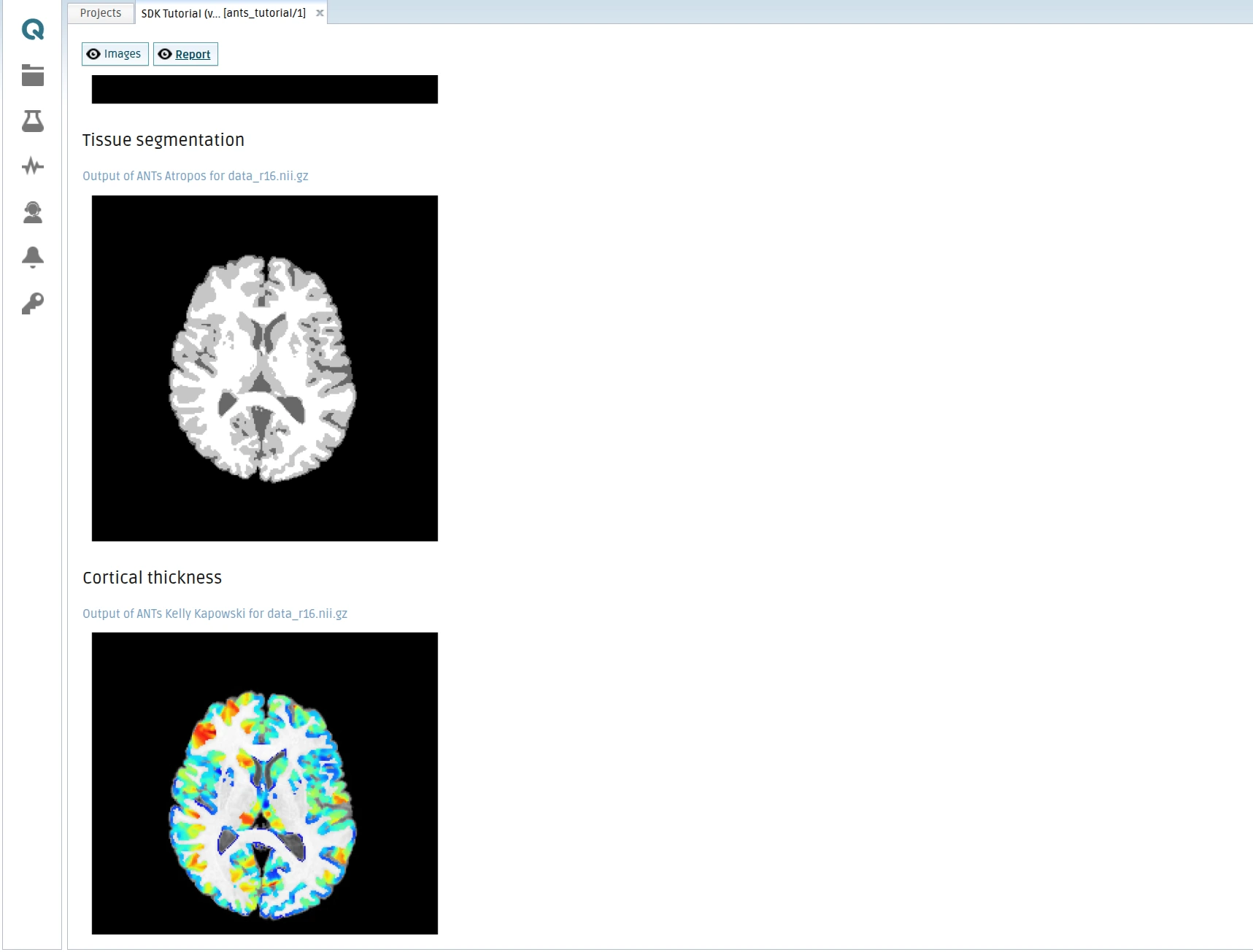
In this extension of the ANTs tutorial, we will focus on reporting and visualizing the different processing steps performed by the tool. Specifically, the tool will generate 2D slices from the processed data, convert them into PNG images, and include them in a report that can be viewed online within the QMENTA Platform.
To achieve this, the tool will create the PNG files locally, upload them as output artifacts, and generate an HTML report that embeds the images to illustrate the intermediate and final results of the analysis. This report will then be uploaded to the platform, where it can be accessed directly from the analysis interface.
The configuration of the tool outputs will be covered in detail in the next section. For now, the focus is on producing the necessary PNG images, uploading them, and assembling the HTML report that references these images correctly:
# ============================================================
# REPORTING PHASE (ALL PLOTTING + BODY_TEMPLATE)
# ============================================================
logger.info("Generating report content")
body_content = ""
for item in report_items:
if "overlay" in item:
item["base_image"].plot(
overlay=item["overlay"],
overlay_cmap="jet",
filename=item["image"]
)
else:
ants.plot(item["source_image"], filename=item["image"])
generated_files.append(item["image"])
body_content += BODY_TEMPLATE.format(
header=item["header"],
src_image=item["image"],
image_description=item["description"],
image_caption=item["description"],
)
report_file = None
if body_content:
report_file = "online_report.html"
with open(report_file, "w") as f:
f.write(HTML_TEMPLATE.format(body=body_content))
generated_files.append(report_file)
We are going to upload the generated results into the QMENTA Platform:
# ============================================================
# UPLOADING PHASE
# ============================================================
logger.info("Uploading outputs to QMENTA Platform")
# Upload original input image for reference
context.upload_file(
source_file_path=fname1,
destination_path="input_image.nii.gz",
modality=fname1_handler.get_file_modality(),
tags=fname1_handler.get_file_tags(),
)
# Upload all generated outputs
for filename in generated_files:
context.upload_file(filename, filename)
# this will report in the QMENTA Platform interface that the tool has completed
context.set_progress(value=100, message="Processing completed")
logger.info("Tool execution finished successfully")
That is all you need to have the run() method working.
Output definition and configuration
Next, we will optionally adapt the tools_output() method to display the results directly in the QMENTA Platform. This method is executed Note that not all outputs are always generated, since the user may choose to execute only a subset of the processing steps. If the QMENTA Platform cannot find the expected files in the output container, the results view will not be displayed. This step is therefore optional but recommended for a more complete and user-friendly presentation of the tool outputs:
def tool_outputs(self):
# Main object to create the results configuration object.
result_conf = ResultsConfiguration()
# Add the tools to visualize files using the function add_visualization
# Online 3D volume viewer: visualize DICOM or NIfTI files.
papaya_1 = PapayaViewer(title="T1 ANTs segmentation and cortical thickness", width="50%", region=Region.center)
# the first viewer's region is defined as center
# Add as many layers as you want, they are going to be loaded in the order that you add them.
papaya_1.add_file(file="input_image.nii.gz", coloring=Coloring.grayscale) # base iamge, using the file name
papaya_1.add_file(file="atropos_processed.nii.gz", coloring=Coloring.custom) # overlay 1, custom coloring shows different colors for different label values.
papaya_1.add_file(file="thickness_processed.nii.gz", coloring=Coloring.hot_n_cold) # overlay 2, hot/cold coloring
# Add the papaya element as a visualization in the results configuration object.
result_conf.add_visualization(new_element=papaya_1)
papaya_2 = PapayaViewer(title="T1 ANTs registration", width="50%", region=Region.right)
# the second viewer's `region` is defined as right. This is required by the Split() element below, which has the property
# orientation set to `vertical`. If it is set to horizontal, then you can choose between 'Region.top' or 'Region.bottom'.
# You can use modality or tag of the output file to select the file to be shown in the viewer.
papaya_2.add_file(file="input_image.nii.gz", coloring=Coloring.grayscale) # using the file modality
papaya_2.add_file(file="warped.nii.gz", coloring=Coloring.red) # using file tag
result_conf.add_visualization(new_element=papaya_2)
# To create a split, specify which ones are the objects to be shown in the split
split_1 = Split(
orientation=OrientationLayout.vertical, children=[papaya_1, papaya_2], button_label="Images"
)
# The button label is defined because this element goes into a Tab element. The tab's "button_label" property
# is a label that will appear to select between different viewer elements in the platform.
html_online = HtmlInject(width="100%", region=Region.center, button_label="Report")
html_online.add_html(file="online_report.html")
result_conf.add_visualization(new_element=html_online)
# Remember to add the button_label in the child objects of the tab.
tab_1 = Tab(children=[split_1, html_online])
# Call the function generate_results_configuration_file to create the final object that will be saved in the
# tool path
result_conf.generate_results_configuration_file(
build_screen=tab_1, tool_path=self.tool_path, testing_configuration=False
)
At the end of the file, you will see this lines:
def run(context):
QmentaSDKToolMakerTutorial().tool_outputs()
QmentaSDKToolMakerTutorial().run(context)
The calling to tools_outputs() is done when running the local test and it will generate the file results_configuration.json. Depending where you run the tool, the run(context) will be called by the local test or the QMENTA Platform execution system when deployed to run the python script. You will learn how to test locally in the next section.
Test the tool locally
At this point you can test the Python program locally.
In order to do so, you will need to install the required dependencies on your Python installation (e.g. system-wide installation, Python environment manager such as virtualenv or pyenv, conda, etc.):
python -m pip install antspyx==0.6.1 qmenta-sdk-lib==2.1
You will need to add the input files in the local_tools/qmenta_sdk_tool_maker_tutorial/local/test/sample_data directory. And then you will need to change the file local_tools/qmenta_sdk_tool_maker_tutorial/local/test/test_tool.py to adapt it to you use case. Replace the file paths in the test:
in_args={
"test_name": inspect.getframeinfo(inspect.currentframe()).function, # returns function name
"sample_data_folder": "sample_data", # Name of the folder where the test data is stored
"input_images": {
"files": [
# Relative paths to the input data stored in the sample_data folder
TestFileInput(
path="data_r16.nii.gz",
file_filter_condition_name="c_image1",
modality=Modality.T1,
tags=[Tag("r16")],
mandatory=1
),
TestFileInput(
path="data_r64.nii.gz",
file_filter_condition_name="c_image2",
modality=Modality.T1,
tags=[Tag("r64")],
mandatory=1
)
],
"mandatory": 1,
},
"perform_steps": [
"do_registration", "do_biasfieldcorrection", "do_thickness"
]
},
Once those requirement are fullfilled, run the test using the pytest module from the folder where local_tools/ is found:
pytest /path/to/local/test/test_tool.py::TestTool
In the following section (Package your tool in a Docker image) you will be able to test the tool packaged in a Docker image, an scenario that represents much more faithfully the execution environment in which the tool will be executed when launched through the QMENTA Platform.
Package your tool in a Docker image
There is another test in the same file to execute the tool inside a container. The Dockerfile can be found here: qmenta-sdk-tool-tutorial/local_tools/ants_tool_maker_tutorial/local/Dockerfile. You will see it’s based on an ubuntu image, but it does not need to be, you can use a lighter base image as long as it supports the code being executed.
That same test will create when run the Docker container for this tool:
pytest /path/to/local/test/test_tool.py::TestToolDocker
Locate the run docker image in your system. In our case, it should be tool_maker_tutorial. After having found it, use it to tag it properly with your namespace:
docker tag tool_maker_tutorial:1.0 {namespace}/tool_maker_tutorial:1.0
Where {namespace} is the name of the Docker namespace, which should coincide with your DockerHub account username, tool_maker_tutorial is the name of the image repository (tool) and 1.0 is the version tag (tool version).
Once you have validated that the tool completes successfully, the next step is to register the image in DockerHub. To do so, you will have to log in first:
docker login
At this point you will be prompted for your username and password. Once authenticated, you will be able to push the image to the Docker registry:
docker push {namespace}/tool_maker_tutorial:1.0
For more information about working with Docker images see Working with Docker images.
Tip
A list of QMENTA’s public Docker images can be found at the qmentasdk Docker registry.
Add the tool to the QMENTA Platform
To follow this step you will need to have an account in QMENTA Platform and have the ability to deploy tools enabled.
Note
Contact us at support@qmenta.com in order to grant you developer privileges to add tools to the QMENTA Platform.
Log in to the QMENTA Platform and access the My Analysis menu. Then, click on Other Options and then My tools.
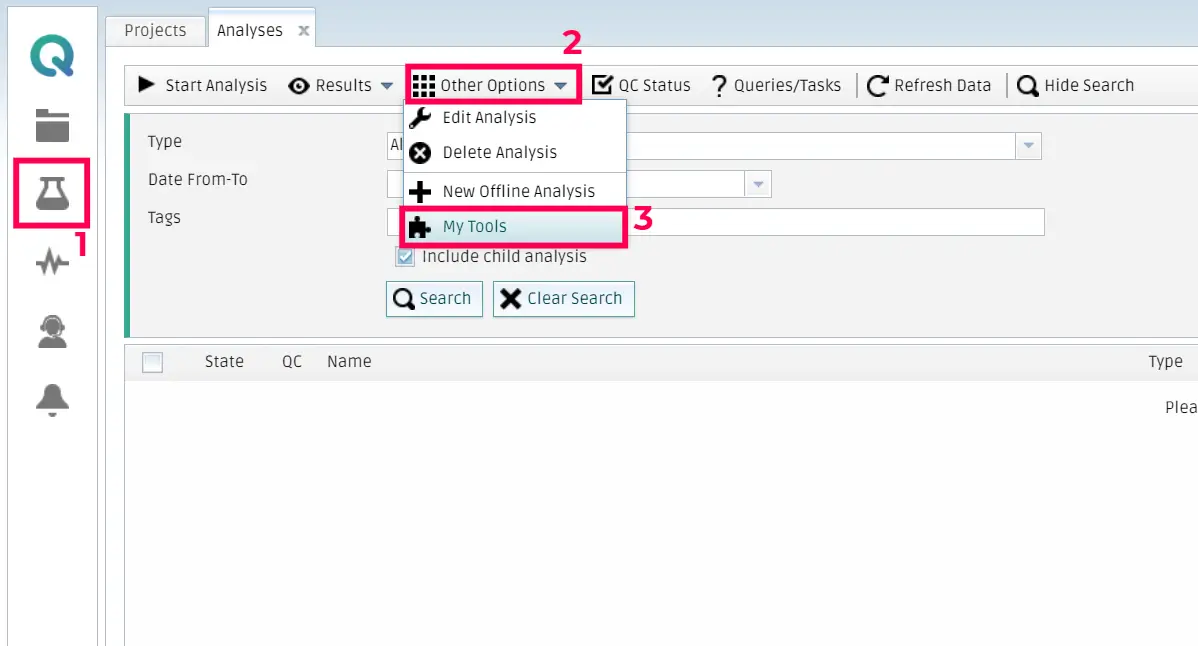
Once the form is visible, you should click on New tool.
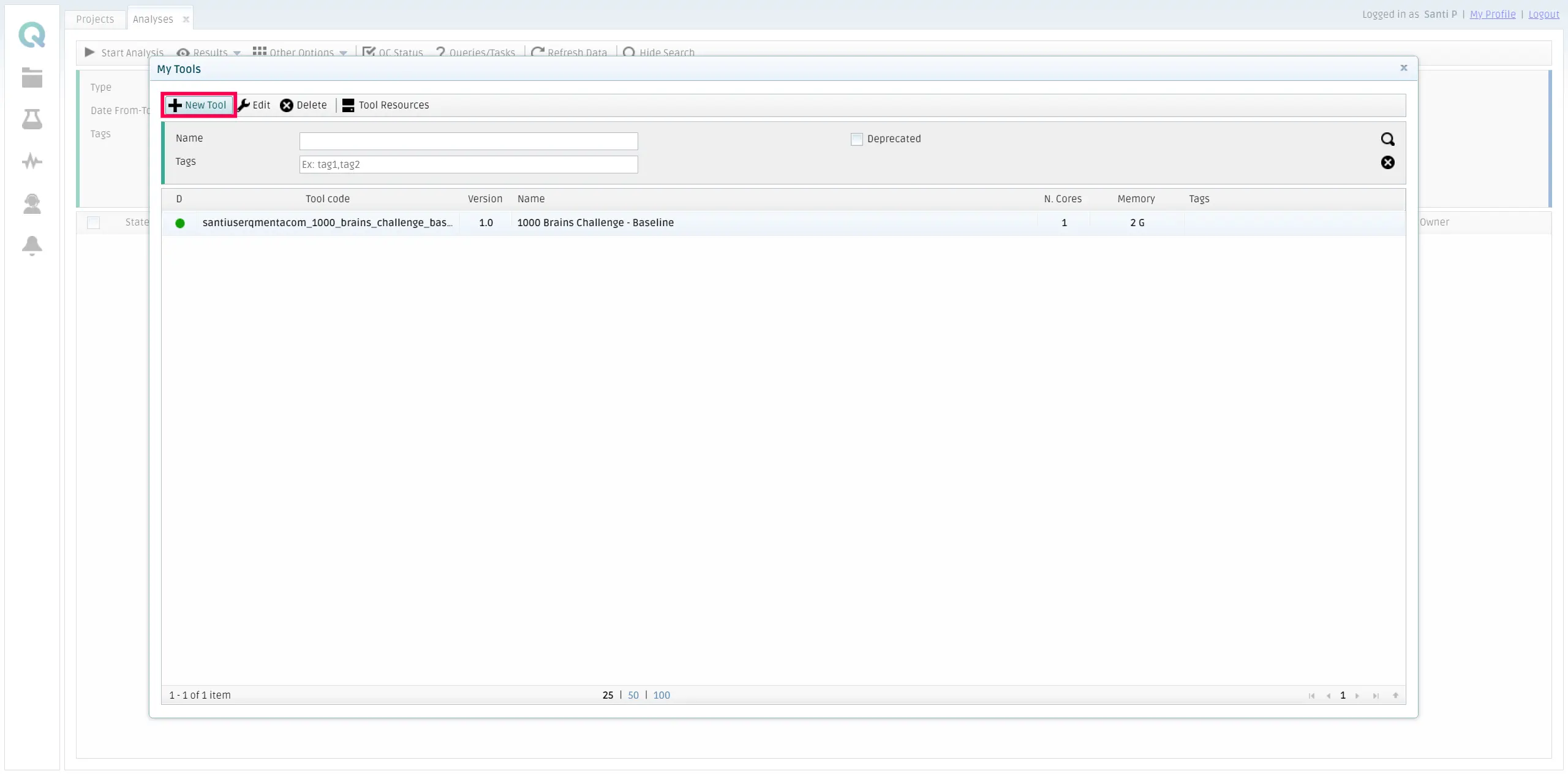
Fill all the fields with the information about your tool. Bear in mind that here the chosen {namespace} is _qmentasdk_, but you should use your own, as explained in Package your tool in a Docker image.
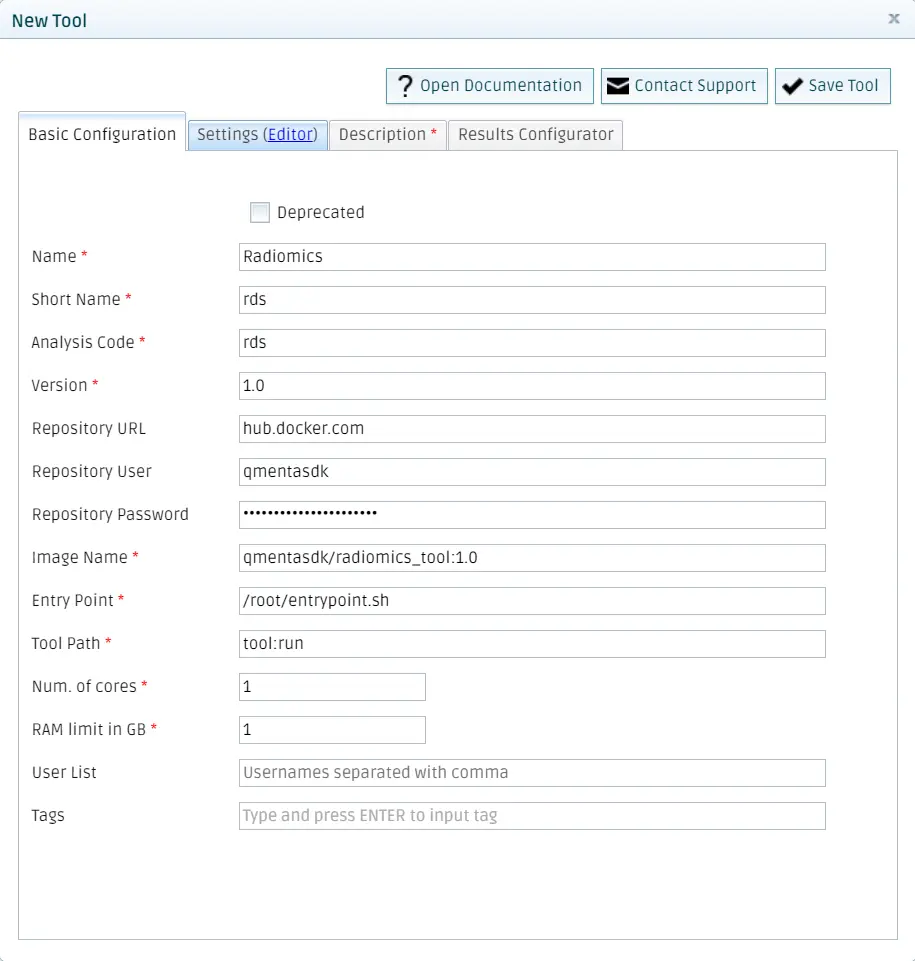
Add settings
Go to the settings tab and click on Editor.
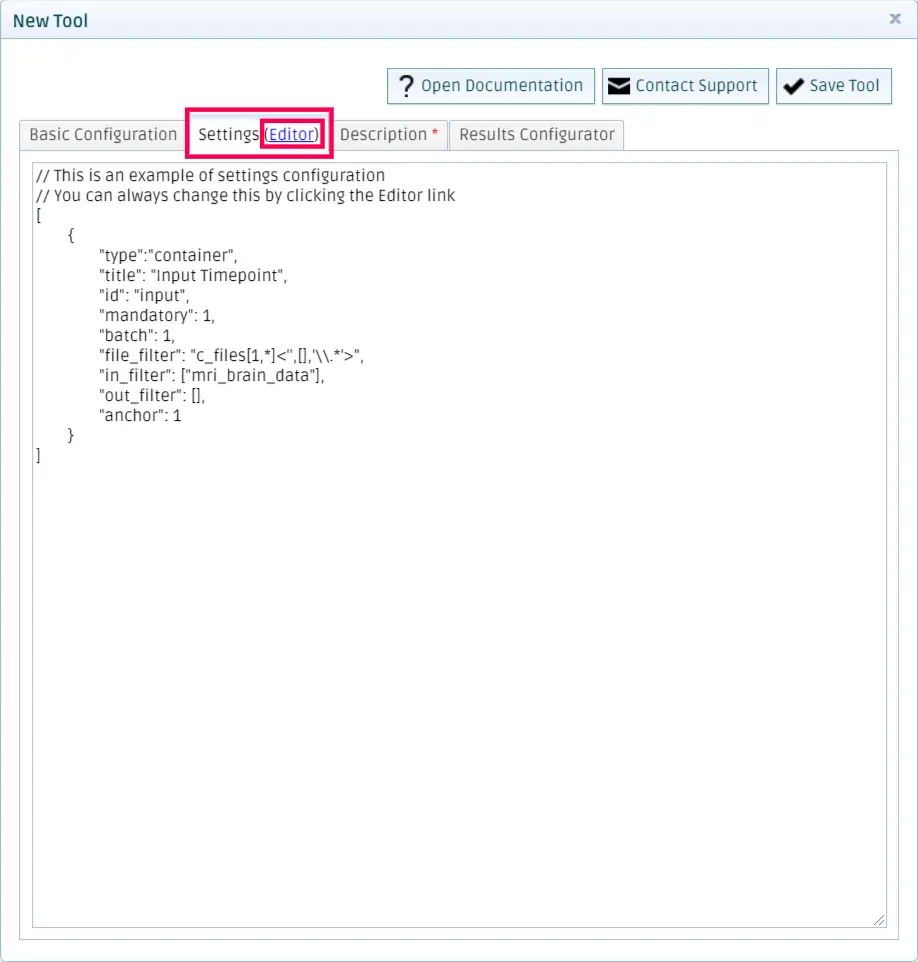
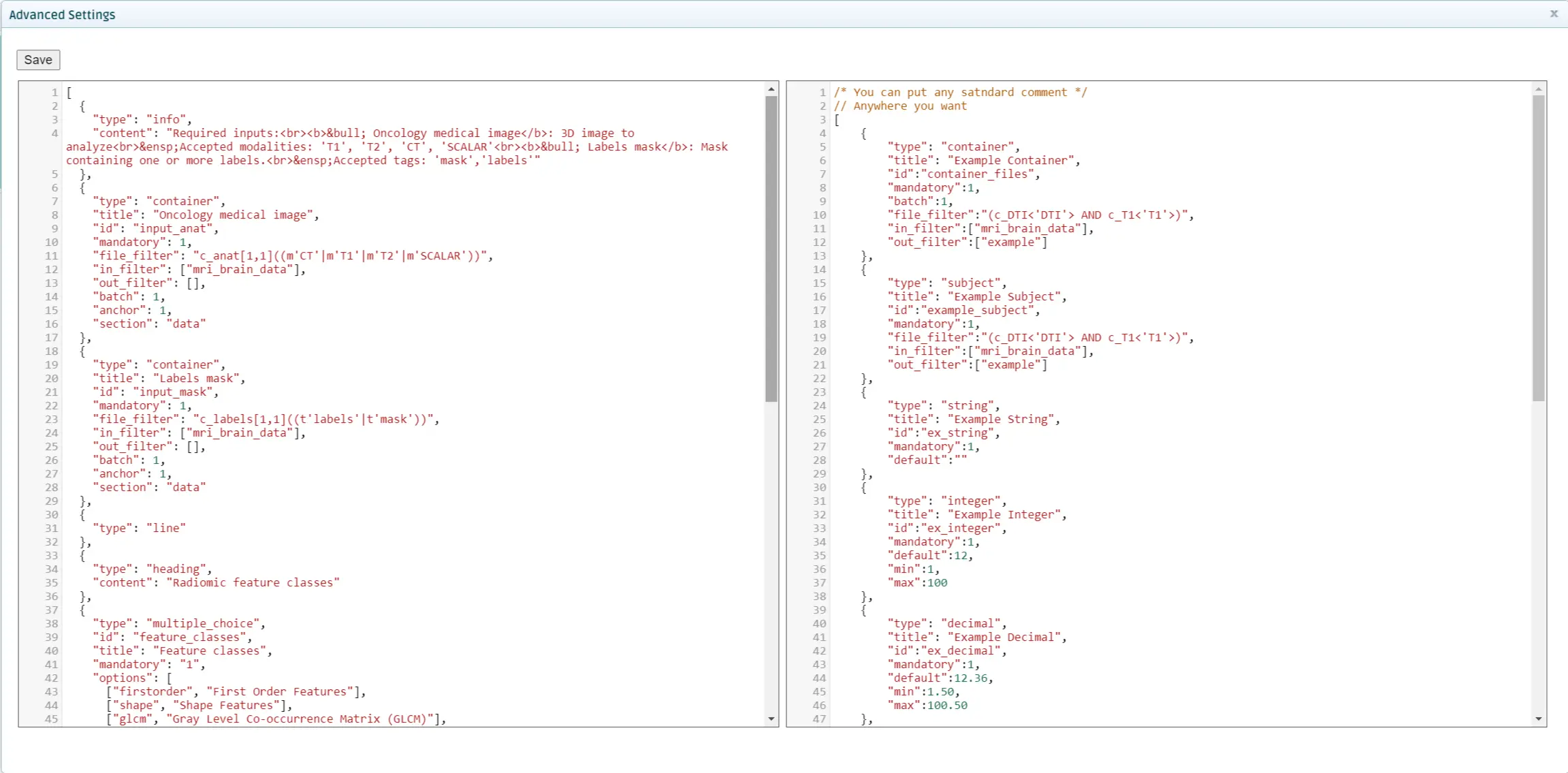
You should copy the settings specified in the settings.json file that you created in Test the tool locally on the left space. You can see some sample settings on the right side for reference.
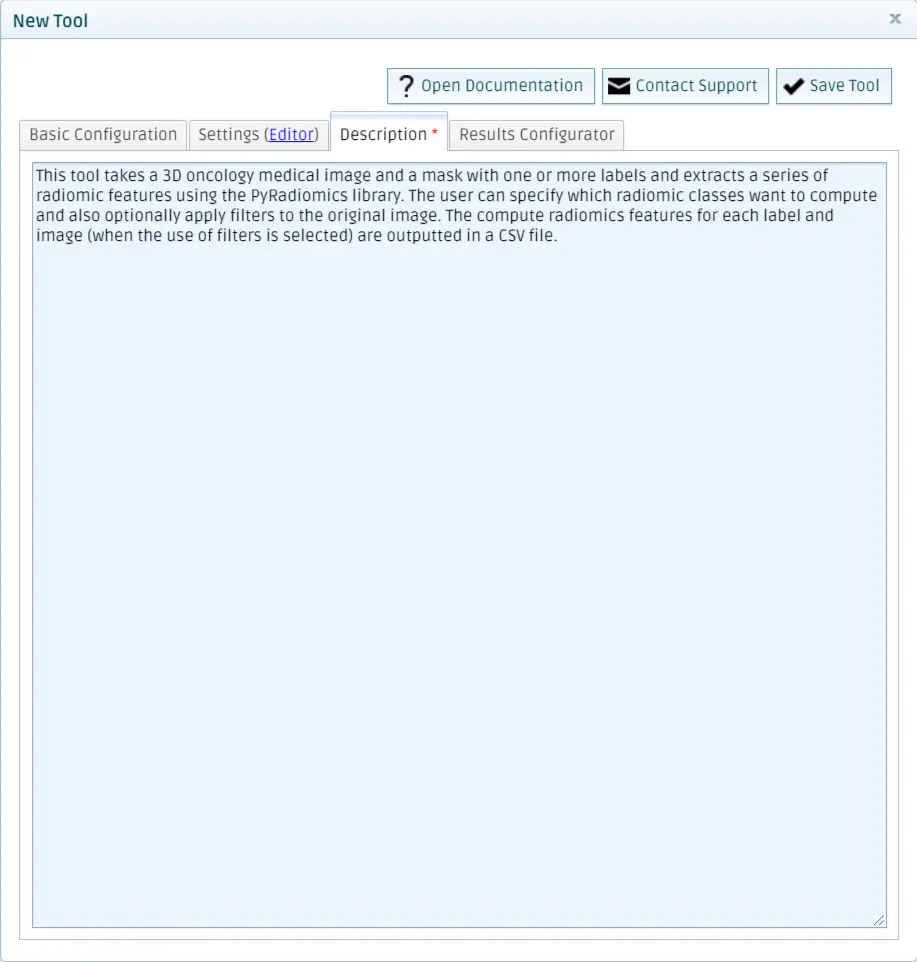
Add a description
This texts is going to be displayed when running the tool from the QMENTA Platform.
Add Results Configuration
Add the configuration to display the results in the QMENTA Platform. When running the tool locally, the file results_configuration.json has been generated. Copy the content of the “screen” key of the JSON into the “Screen layout” text box. You can add widgets to visualize results by selecting them on the dropdown list and by clicking “Add”. Add two Papaya viewers and an “HTML Inject” widget, and they should appear listed. Click on the Config link next to each widget to add the files that you want to present there, they should also be in the results_configuration.json file, in the “tools” part.
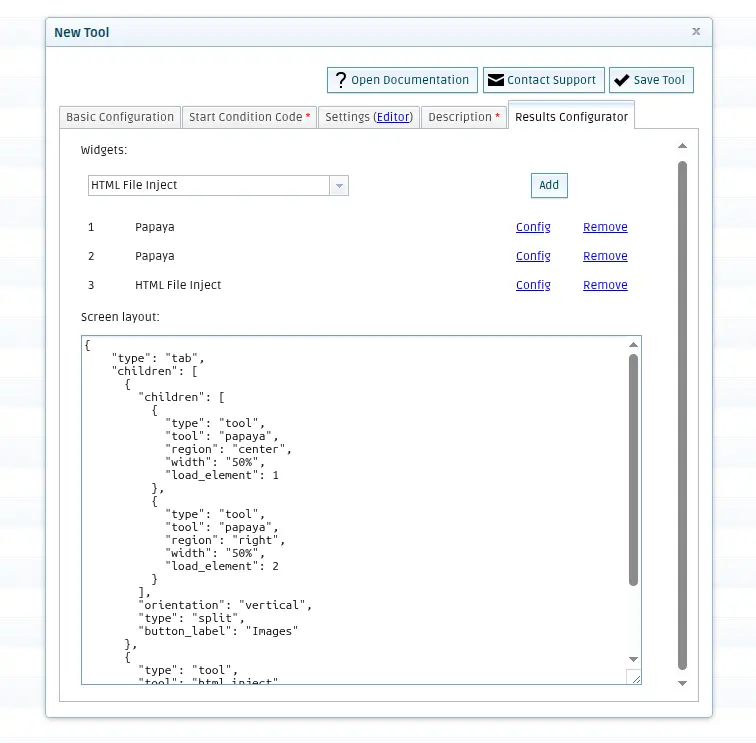
Finally, click on Save Tool. The QMENTA Platform will pop-up a success message, and after closing the form to add tools, you will see your newly created tool in your My Tools section.
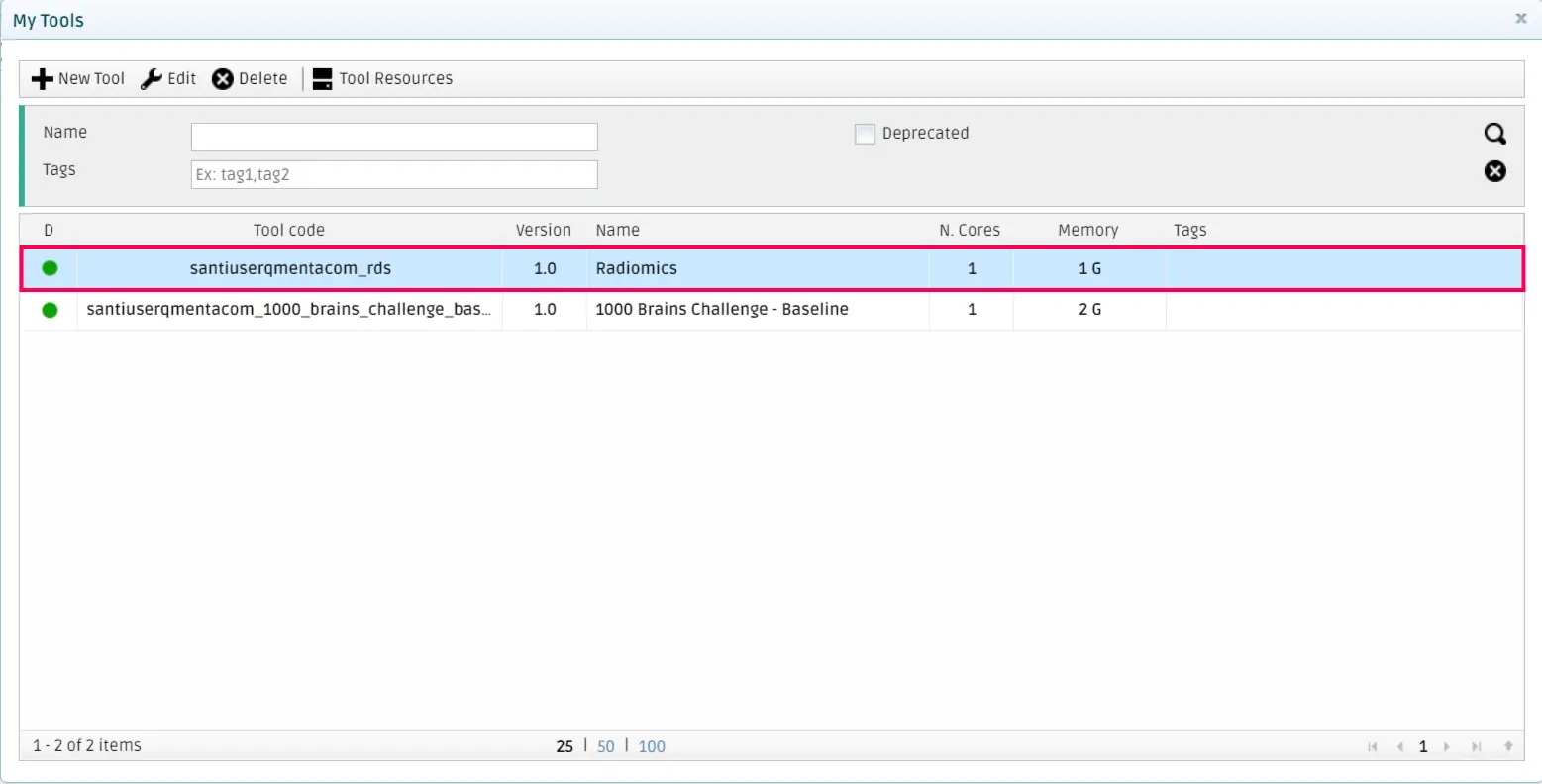
Note
As you will have noticed, your username is preprended to the tool_code you specify. This is done so that each tool is unique within the whole QMENTA Platform, so you don’t have to worry about having unique names for your tools.
Run your tool in the QMENTA Platform
After the successful completion of the previous steps, the tool is ready to be run in the QMENTA Platform.
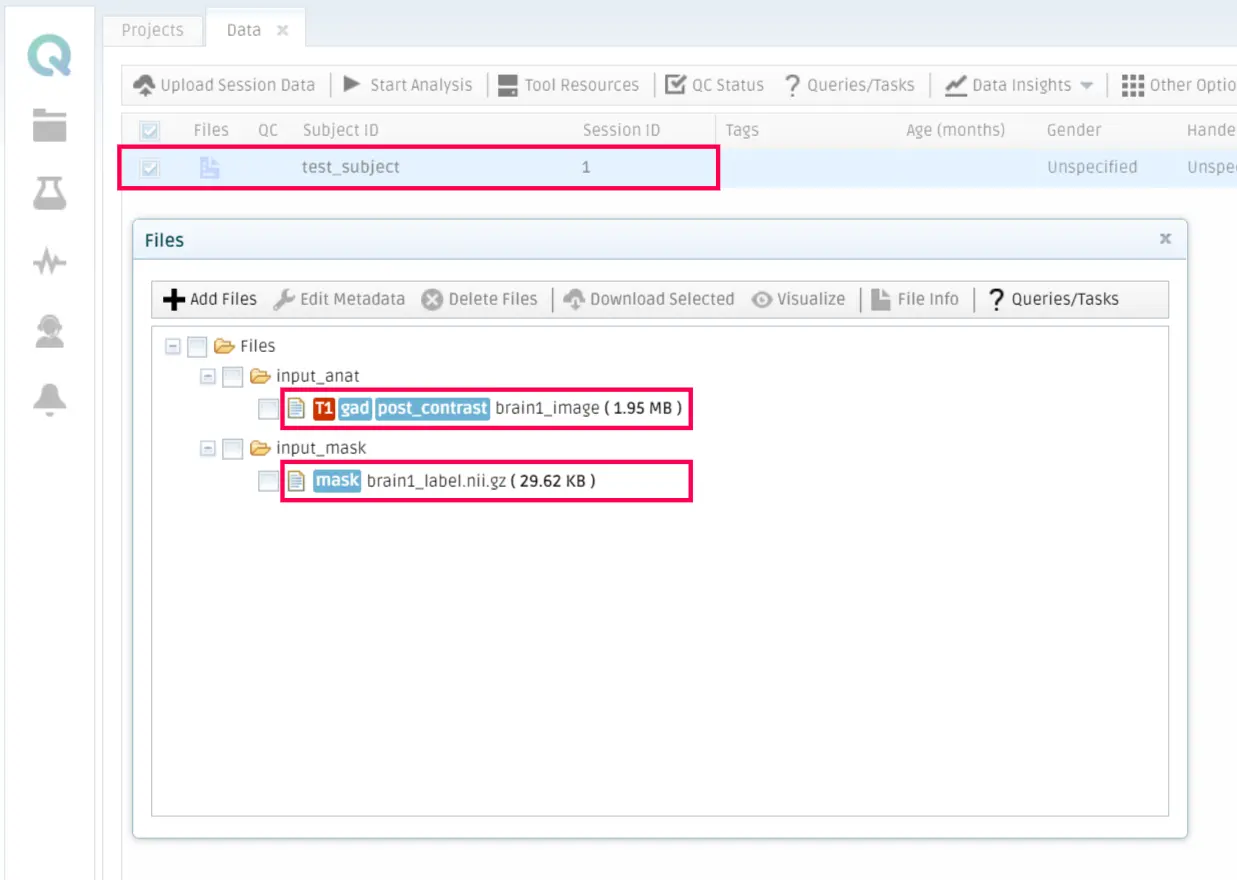
To do so, you should go to My Data in the QMENTA Platform, select a subject that contains the necessary files to run the tool (properly tagged) and click on Start Analysis.
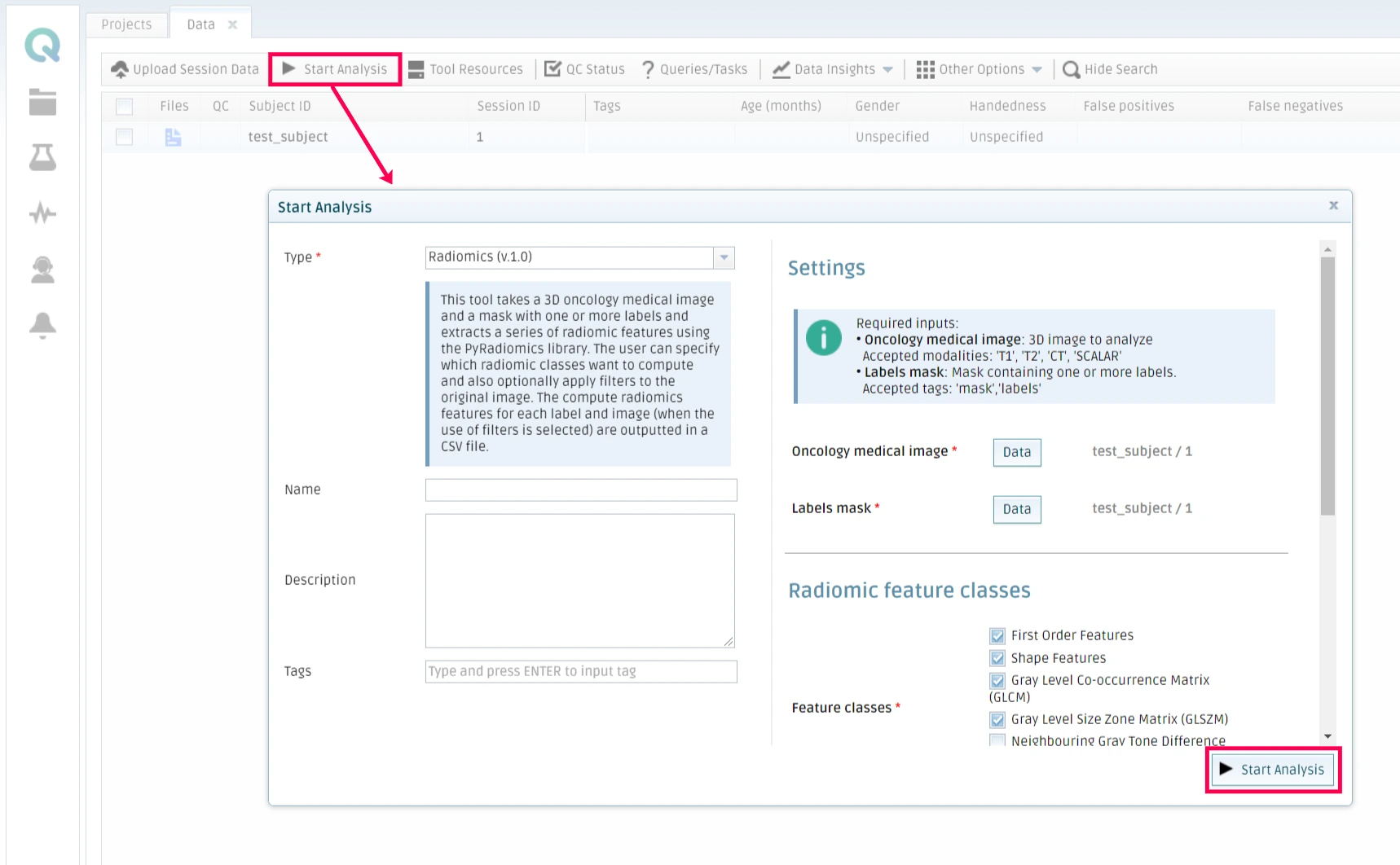
Then select your tool, choose the parameters you want it to run with and optionally add a name and a description to the analysis. Click on Start Analysis when the configuration is ready.
The QMENTA Platform will run the analysis and send an email whenever it finishes.
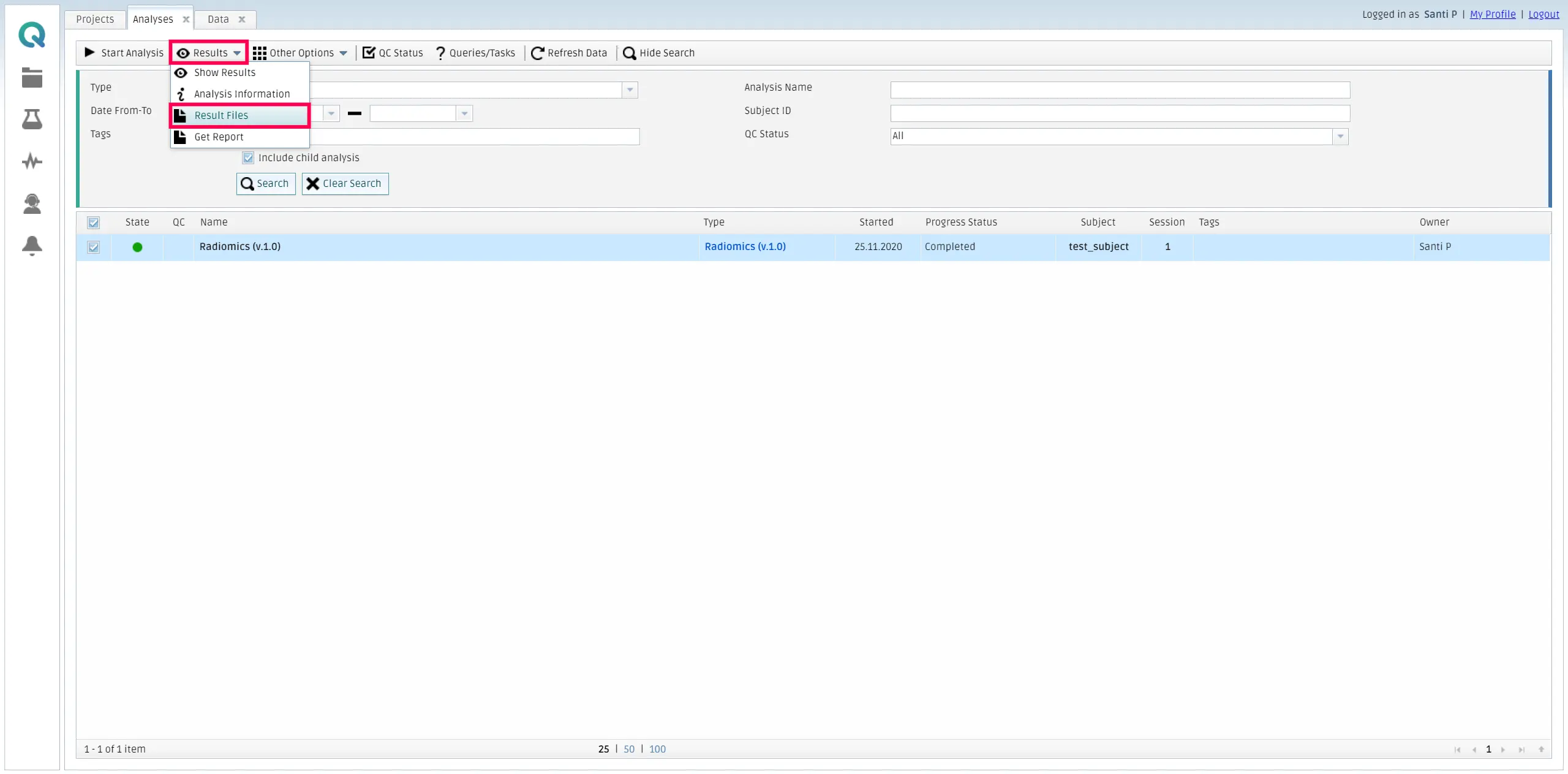
Then, you can retrieve the results by going to My Analysis, selecting the finished analysis, and then click on Results and Result files inside the Results tab.
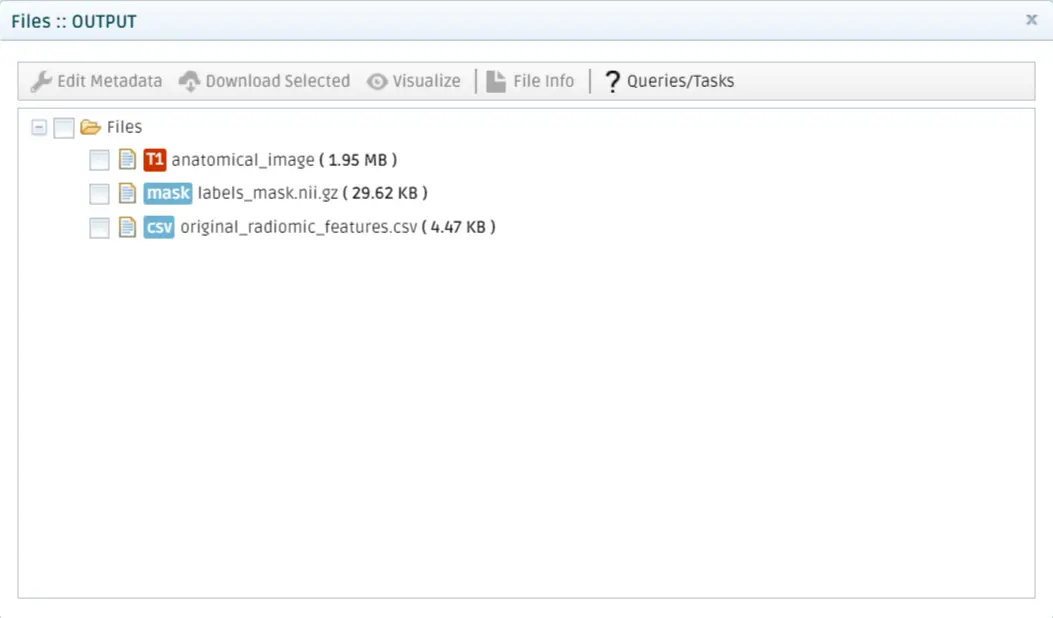
Click on Show Results in the Results dropdown menu to visualize the results. The QMENTA Platform will use the configuration that you set in the Result Configuration section of the tool to render the results using the Papaya viewer and display the HTML file set in the configuration to be displayed. Switch views by clicking the buttons on the top left corner.
Papaya viewer with the segmentation and reistration results (Images):
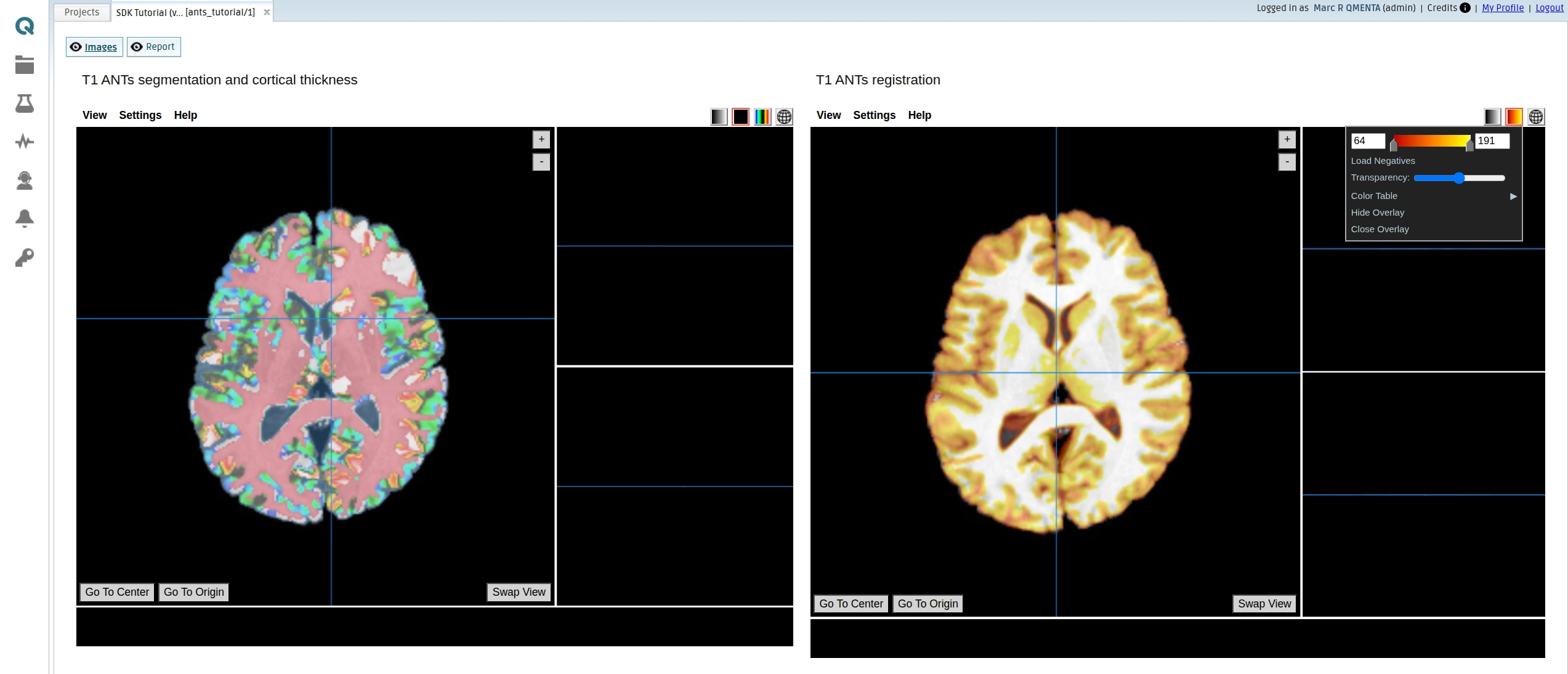
HTML report rendered with the different images showing the processed data (Report):